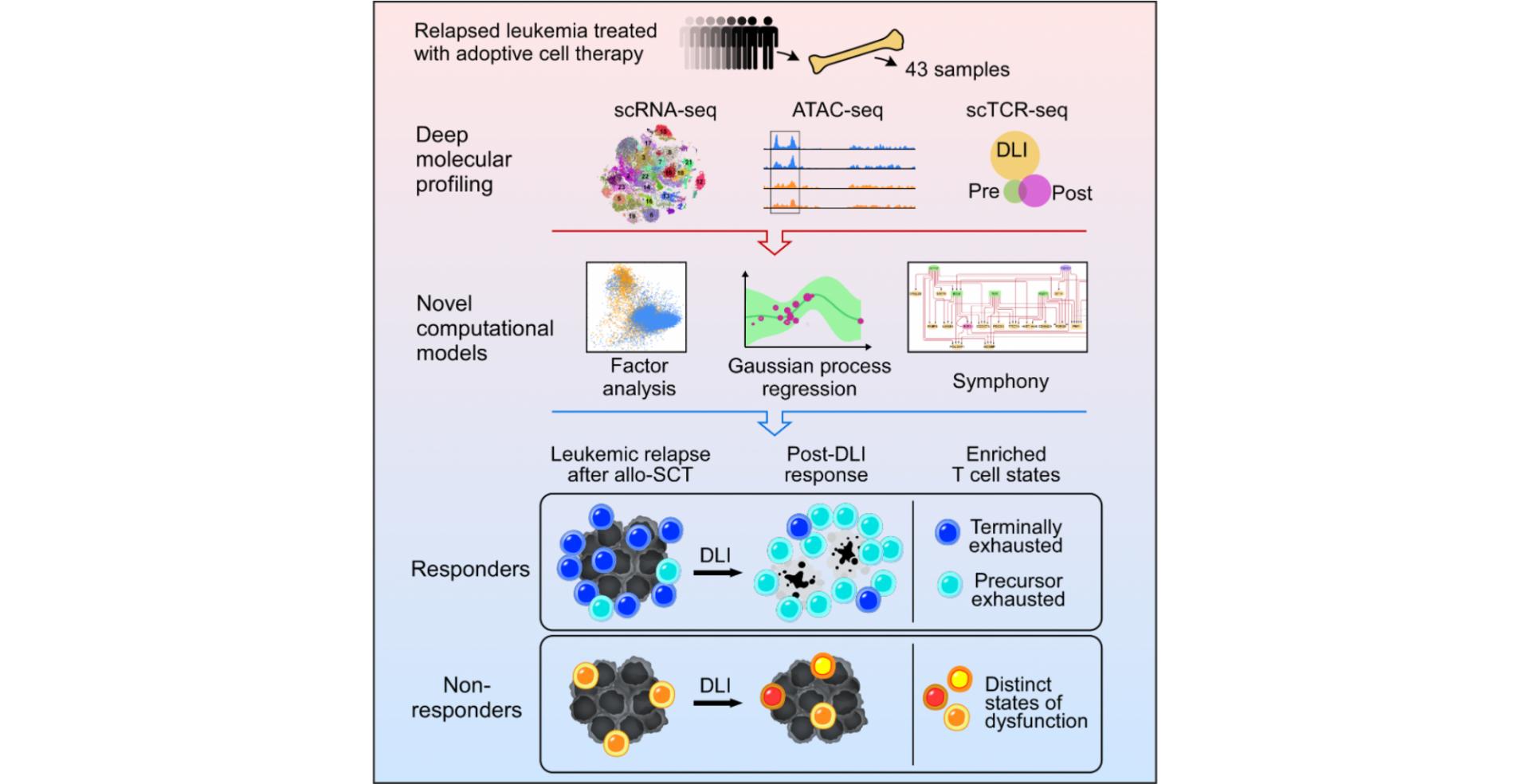
A new study co-authored by Pavan Bachireddy, MD (Assistant Professor, Division of Cancer Medicine at the University of Texas MD Anderson Cancer Center) and Elham Azizi, PhD (Assistant Professor, Department of Biomedical Engineering and Irving Institute for Cancer Dynamics) was recently published in Cell Reports and revealed the cellular state dynamics in the tumor microenvironment during response and resistance to adoptive cellular therapy.
Donor lymphocyte infusion or DLI, a form of adoptive cellular immunotherapy for bone marrow transplant patients in which lymphocytes from the donor are transferred to the recipient, is commonly used for patients with relapsed leukemia following allogeneic hematopoietic stem cell transplant. However, the underlying mechanisms that make DLI an efficient treatment are poorly understood. To elucidate these mechanisms, Bachireddy, Azizi et al. developed innovative computational tools to analyze over 90,000 single-cell T cell transcriptomes, bulk chromatin accessibility profiles, and single T cell clonality data from bone marrow biopsies of a cohort of patients treated with DLI after relapsed chronic myelogenous leukemia. Specifically, Dr. Azizi led the development of two machine learning methods: a hierarchical Gaussian Process model for uncovering temporal dynamics from longitudinal single-cell data, and Symphony for inferring gene regulation from integration of single-cell transcriptomic and epigenetic data. Applying these methods revealed specific subsets of exhausted T cells (terminal and precursor exhausted T cells) and their regulatory circuitry that define the response or resistance to DLI therapy in humans. The authors proposed that the developed computational framework could be further used to analyze longitudinal single-cell data sets for other cancer types and other diseases beyond oncology.
Read the article here.